Using Label Clustering to Cluster Data
The Label Clustering task performs a clustering process according to the k-means approach after having aggregated and filtered data according to a subset of label variables.
The output of the task is a collection of clusters characterized by:
a (positive integer) index,
a central vector (centroid) and
a dispersion value measuring the normalized average distance of cluster members from the centroid.
In the following examples, which illustrates the three phases of the analysis, the list of values assumed by the label variables in a given pattern of the dataset is called tag:
Data grouping: Examples in the training set characterized by the same tag are grouped together and considered as a single representative record. The mean (or median, or medoid, according to the option specified by the user) among the values of each profile attribute is computed and assigned to the corresponding variable in the unified record.
Data filtering: Representative records with profile variables which have undesired properties are discarded. Two filter conditions are presently implemented:
Minimum number q of occurrences: Records that do not derive from a group of at least q patterns of the training set with the same tags are removed as statistically they are not highly representative.
Maximum dispersion coefficient σ: If the values of the profile variables of the group of patterns leading to a representative record present a dispersion coefficient (computed with respect to the desired central value) greater than σ, that record presents an irregular behavior that can deteriorate the results of the clustering procedure, and it is consequently discarded.
Data clustering: A k-means (or k-medians, or k-medoids, according to the option specified by the user) clustering algorithm is employed to aggregate representative records with similar profiles. The centroid of each cluster provides the values of the profile variables to be used in a subsequent Apply Model task when a new pattern is assigned to that cluster.
Label clustering can also be employed for the solution of signal prediction problems, where the behavior of a mono-dimensional output signal (described by the profile variables) has to be predicted starting from a set of label attributes.
Prerequisites
the required datasets have been imported into the process
the data used for the model has been well prepared and the final dataset contains profile and label variables.
To preserve generality a profile attribute can also be a label variable.
If nominal profile attributes are considered, the k-modes variant is adopted to allow their treatment. Optionally, a variable with the cluster id role can be included in the dataset, providing the initial cluster assignment for each pattern.
If a weight attribute is present, its values are employed as a measure of relevance for each example, thus affecting the position of the cluster centroid.
Additional tabs
Along with the Options tab, where the task can be configured, the following additional tabs are provided:
Documentation tab where you can document your task,
Parametric options tab where you can configure process variables instead of fixed values. Parametric equivalents are expressed in italics in this page (PO).
Clusters & Results tabs, where you can see the output of the task computation. See Results table below.
Procedure
Drag and drop the Label Clustering task onto the stage.
Connect a Split Data task, which contains the attributes you want to cluster, to the new task.
Double click the Label Clustering task.
Configure the attributes described in the table below.
Save and compute the task.
Label K-Means Clustering options | ||
Parameter Name | PO | Description |
|---|---|---|
Attributes to consider for clustering | profilenames | Drag and drop the attributes that will be used as profile attributes in the clustering computation. |
Label attributes | labelnames | Drag and drop the attributes that that will be considered as labels in the clustering computation. |
Clustering type | centroidtype | Three different approaches for computing cluster centroids are available:
|
Clustering algorithm | kmeanstype | Three different clustering algorithms are available:
|
Distance method for clustering | distmethod | The method employed for computing distances between examples. Possible methods are: Euclidean, Euclidean (normalized), Manhattan, Manhattan (normalized), Pearson. Details on these methods are provided in the Distance parameter of the Managing Attribute Properties page. |
Distance method for evaluation | evaldistmethod | Select the method required for distance, from the possible values: Euclidean, Euclidean (normalized), Manhattan, Manhattan (normalized), Pearson. For details on these methods see the Managing Attribute Properties page. |
Normalization for ordered variables | normtype | Type of normalization adopted when treating ordered (discrete or continuous) variables. Every attribute can have its own value for this option, which can be set in the Data Manager. Details on these options are provided in the Distance parameter of the Managing Attribute Properties page. These choices are preserved if Attribute is selected in the present menu; every other value (e.g. Normal) supersedes previous selections for the current task. |
Initial assignment for clusters | assigntype | Select the procedure to be adopted for the initial assignment of points to clusters; it may be one of the following:
|
(Optional) attribute for initial cluster assignment | clusteridname | Optionally select a specific attribute from the drop-down list, which will be used as an initial cluster assignment. |
(Optional) attribute for weights | weightname | Optionally select an attribute from the drop-down list, which will be used as a weight in the clustering process. |
Number of clusters to be generated | nclustot | The required number of clusters. The number of clusters cannot exceed the number of different examples in the training set. |
Number of executions | ntimes | Number of subsequent executions of the clustering process (to be used in conjunction with Random as the Initial assignment for clusters option); the best result among them is retained. |
Maximum number of iterations | nkmiter | Maximum number of iterations of the k-means inside each execution of the clustering process. |
Minimum decrease in error value | mindecrease | The error value corresponds to the average distance of each point from the respective centroid. This value, measured at each iteration, should gradually decrease. When the error decrease value (i.e. the difference in error between the current and previous iteration) falls below the threshold specified here, the clustering process stops immediately since it is supposed that no further significant changes in error will occur. |
Minimum number of occurrences | minrepl | Minimum number of examples in the training set that must be characterized by a given tag so that it passes the filtering phase. |
Maximum dispersion coefficient | maxsdev | If the profile attribute values present a dispersion coefficient (computed with respect to the desired central value) greater than the value entered here, the record presents an irregular behavior that can deteriorate the results of the clustering procedure, and is consequently discarded. |
Initialize random generator with seed | initrandom | If checked, the positive integer shown in the box is used as an initial seed for the random generator; with this choice two iterations with the same options produce identical results. |
Keep attribute roles after clustering | keeproles | If selected, roles defined in the clustering task (such as profile, labels, weight and cluster id) will be maintained in subsequent tasks in the process. |
Filter patterns before clustering | filter | If selected, data is filtered, otherwise all the representative records are considered in the clustering process. |
Aggregate data before processing | aggregate | If checked, identical patterns in the training set are considered as a single point in the clustering process. |
Append results | append | Additional attributes produced by previous tasks are maintained at the end of the present one, rather than being overwritten. |
Results
The results of the task can be viewed in three separate tabs:
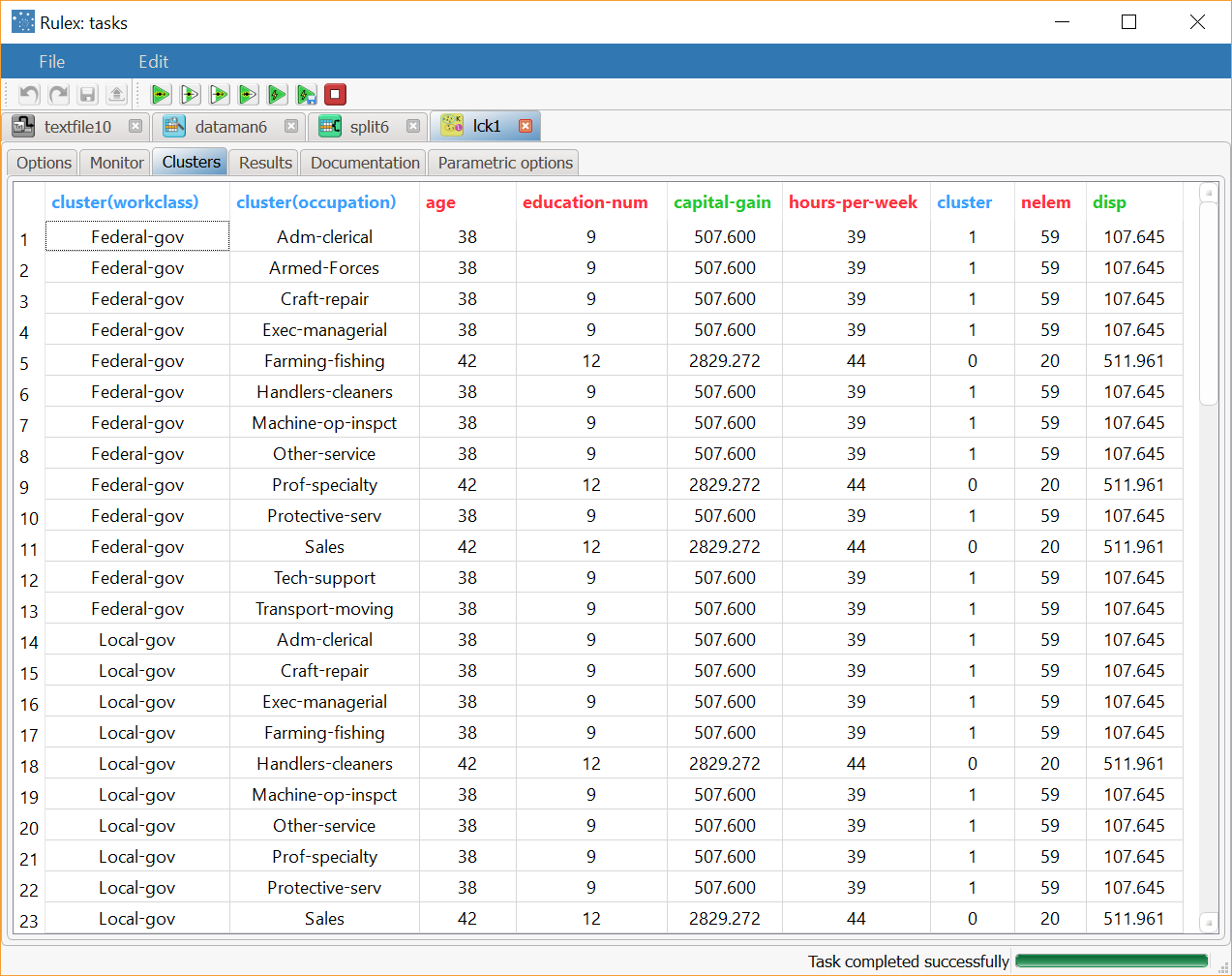
The Clusters tab displays a spreadsheet with the values of the profile attributes for the centroids of created clusters, together with the number of elements and the dispersion coefficient (given by the normalized average distance of cluster members from the centroid) for each of them. In particular, the cluster, nelem and disp columns respectively contain the index of the cluster, the number of elements and the dispersion coefficient. Since several tags may be included in the same cluster, the characteristic values of each cluster may appear in more than one row of the spreadsheet displayed in the Clusters tab. By ordering the column
clusterwe can easily retrieve all the tags belonging to each cluster.
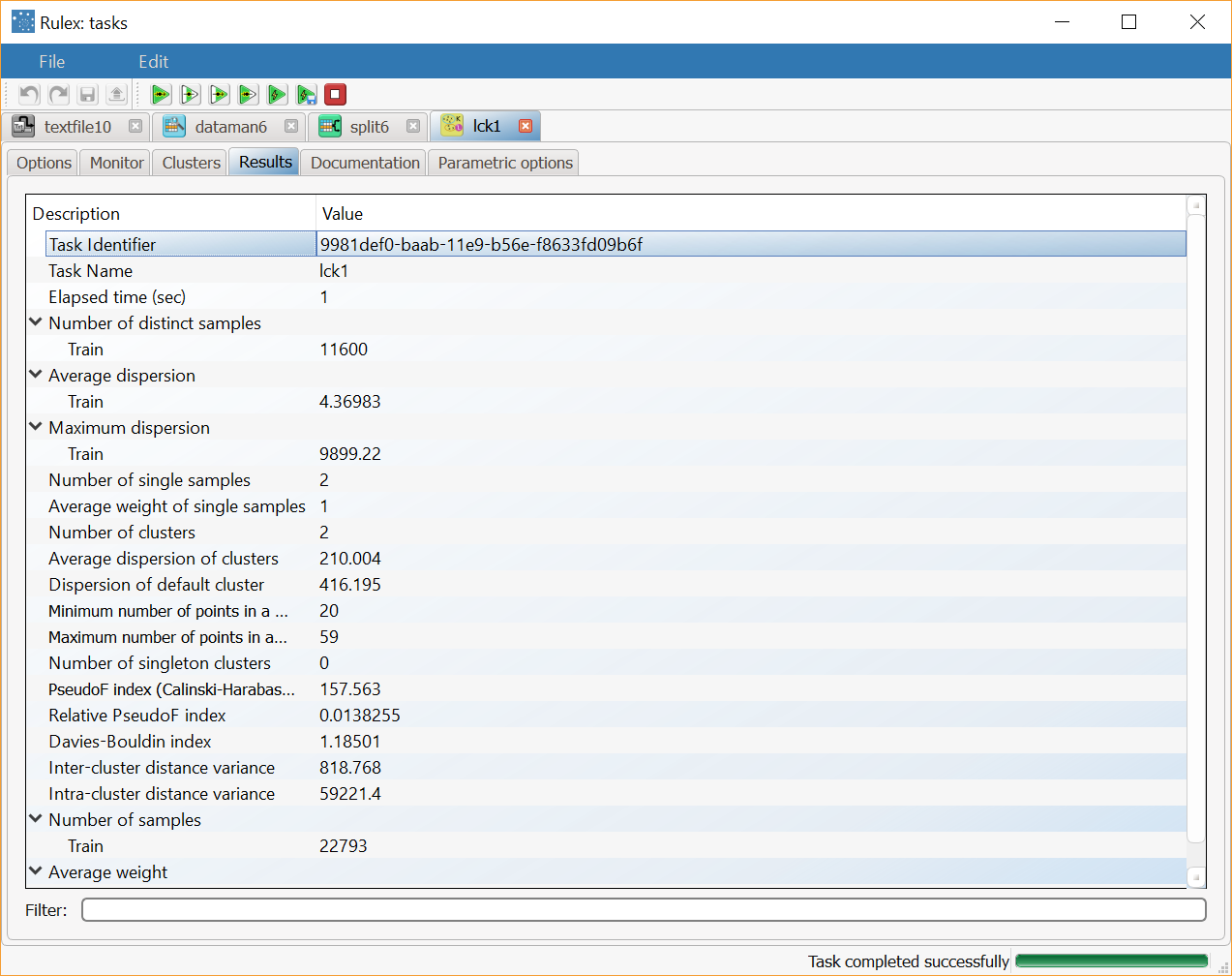
The last row, characterized by a null index in the column cluster, reports the values pertaining to the default cluster, obtained by including all the elements if the training set in a single group. To point out the generality of this special cluster all the values in its tag are set to missing.The Results tab, where a summary on the performed calculation is displayed, among which:
the execution time,
the number of valid training samples,
the average weight of training samples,
the number of distinct tags in the training set,
the average, minimum and maximum dispersion coefficient for these tags,
the number of tags present in only one training sample and their average weight,
the number of clusters built,
the average dispersion of clusters,
the dispersion coefficient of the default cluster,
the minimum and the maximum number of points in clusters,
the number of singleton clusters, including only a point of the training set.
Example
The following examples are based on the Adult dataset.
Scenario data can be found in the Datasets folder in your Rulex installation.
The scenario aims to divide the dataset into a specific number of defined clusters.
The following steps were performed:
First we import the adult dataset with an Import from Text File task.
Define the attributes that need to be ignored in the Data Manager task.
Split the dataset into a test and training set with a Split Data task.
Generate the required clusters in the Label Clustering (K-means) task.
View the results of the Label Clustering task with a Data Manager task.
Apply the rules to the dataset with an Apply Model task.
View the results of the forecast from the Apply Model task using the Take a Look function.
Procedure | Screenshot |
|---|---|
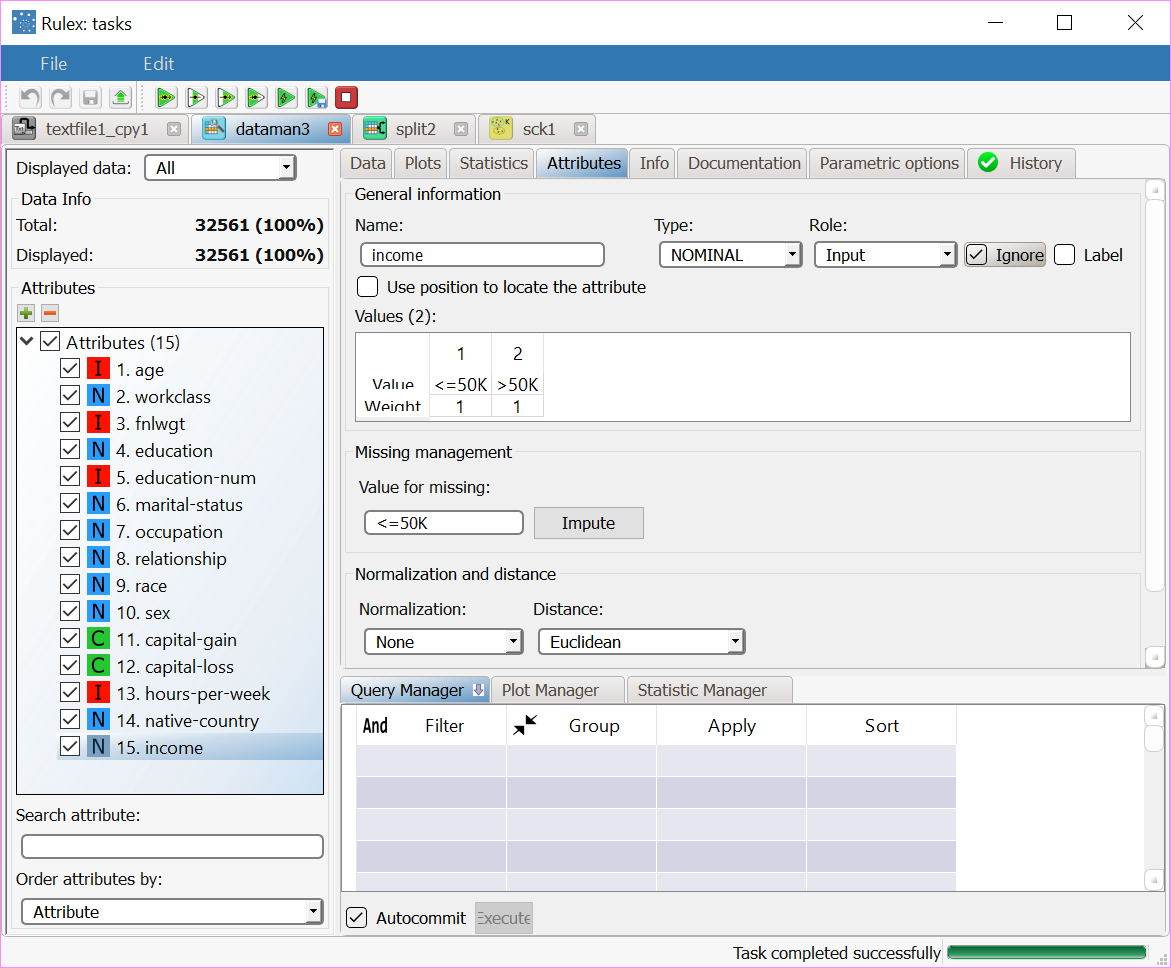
After importing the adult dataset file into the process via the Import from Text file task, double-click the Data Manager task, select the Income attribute and check Ignore in the Attributes tab. Split Data task in the process, and split the dataset as follows:
Save and compute the task. | |
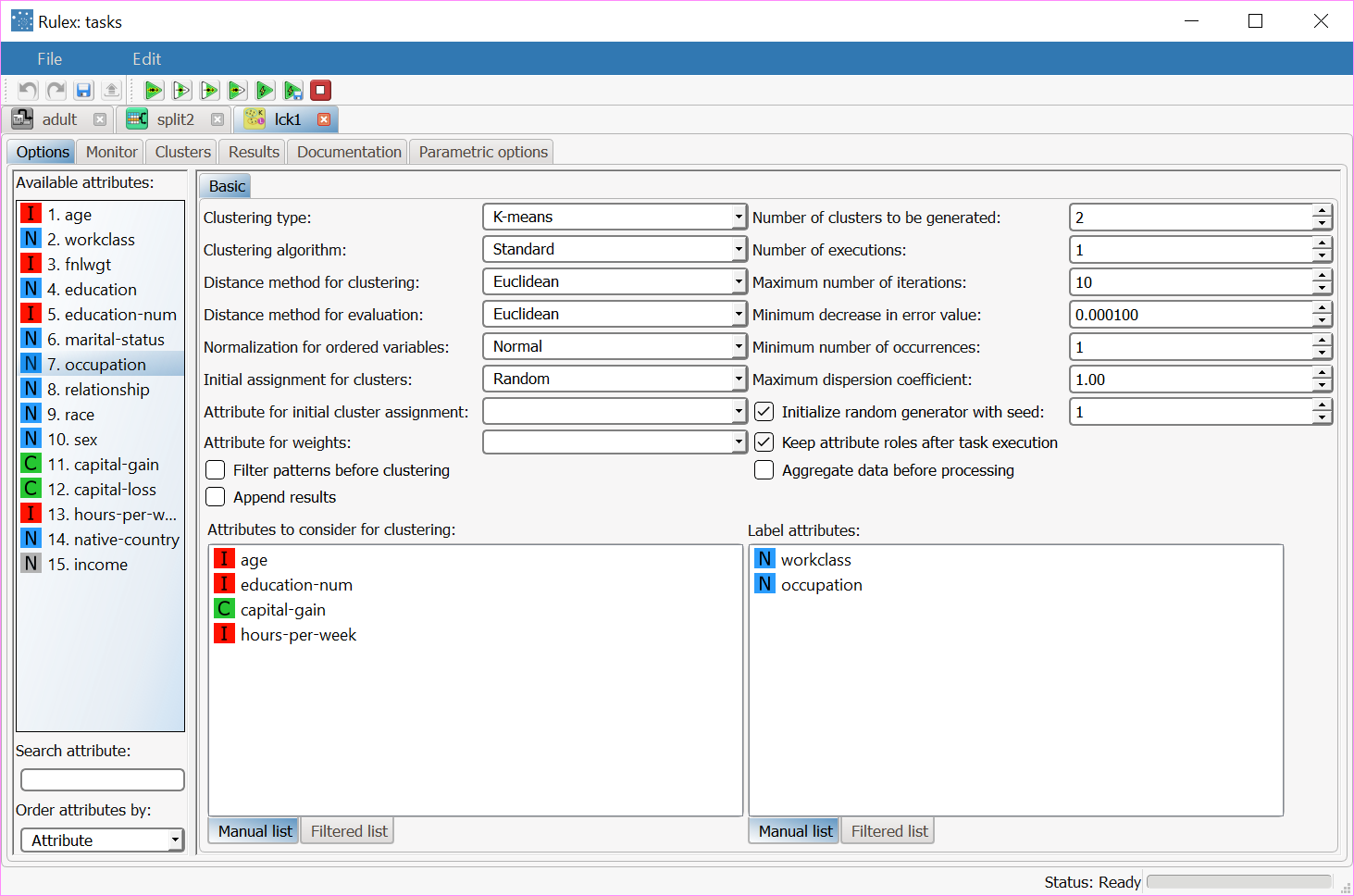
Double-click the Label Clustering (K-means) task, and configure it as follows:
| |
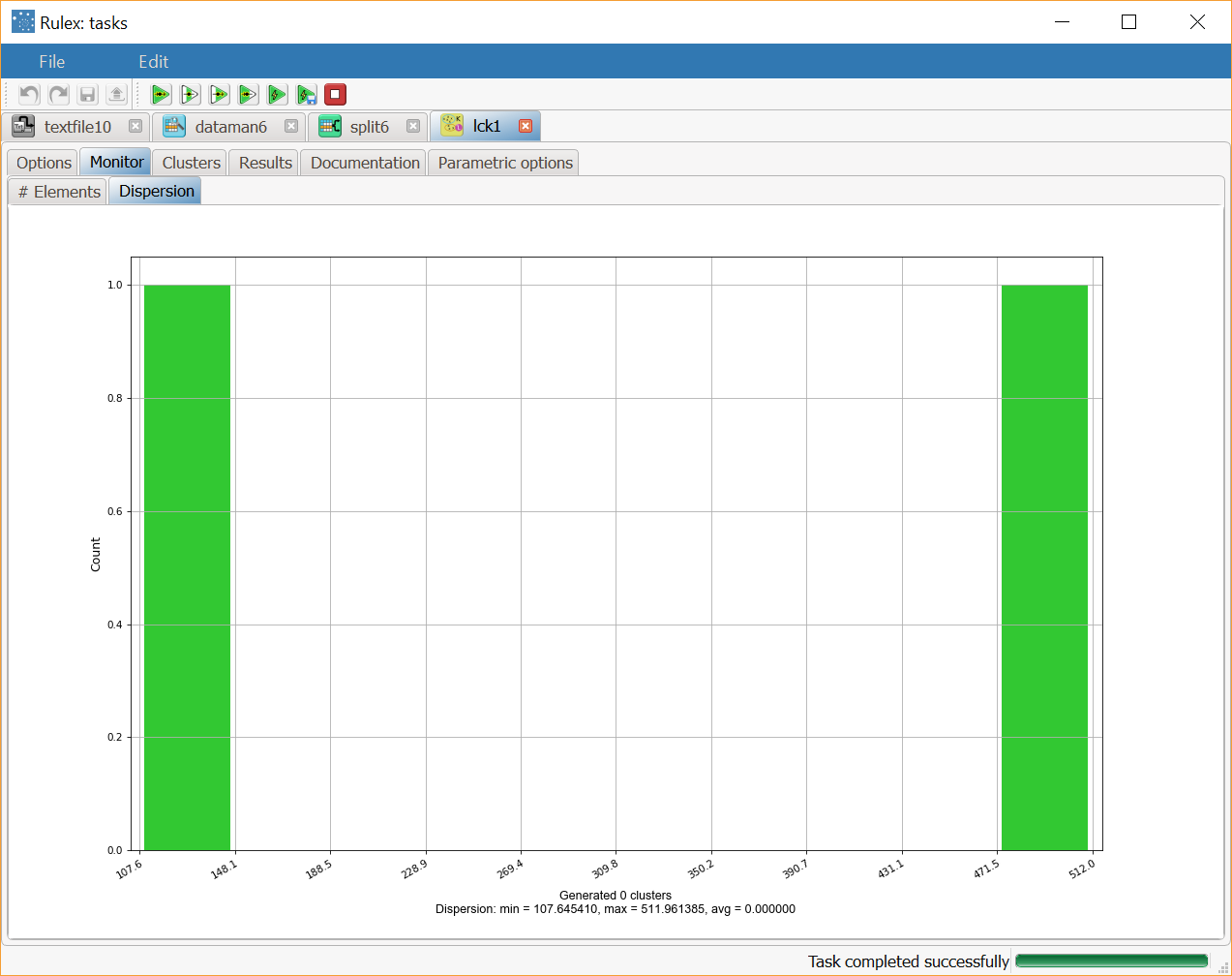
After clicking Compute process to start the analysis, the properties of the generated clusters can be viewed in the Monitor tab of the Standard Clustering task and at the end of the process the dispersion coefficients of the clusters are distributed as in the screenshot. A similar histogram can be viewed for the number of elements, by opening the corresponding #Elements tab. | |
After the execution of Label Clustering task we obtain two clusters whose characteristics are displayed in the Clusters panel of the task. In each row of the spreadsheet the first two fields, called cluster(workclass) and cluster(occupation), refer to a specific tag included in some patterns of the training set, whereas the subsequent 5 columns contain the components of the centroids for the two clusters. The columns cluster, nelem and disp provide the cluster index, the number of elements and the dispersion coefficient, respectively. The last row reports the values characterizing the default cluster, obtained by including all the elements of the training set in a single group. | |
Clicking on the Results tab displays a summary of the computation performed, with:
| |
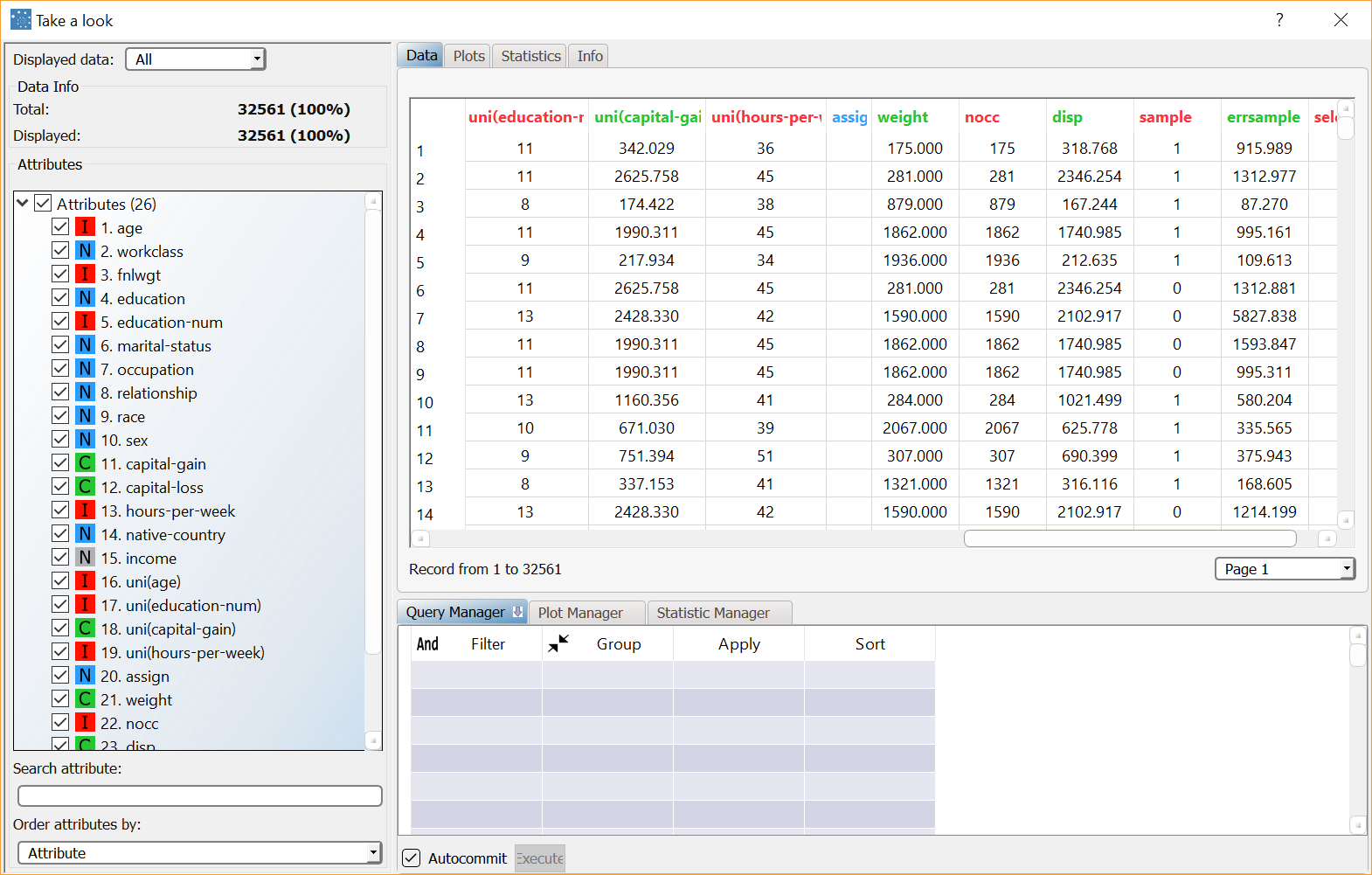
To view information concerning the representative records created by the Label Clustering (K-means) task, right-click the clustering task and select Take a look. Eleven additional result columns have been added:
For each sample (row) of the training set we have the following result variables (columns)
| |
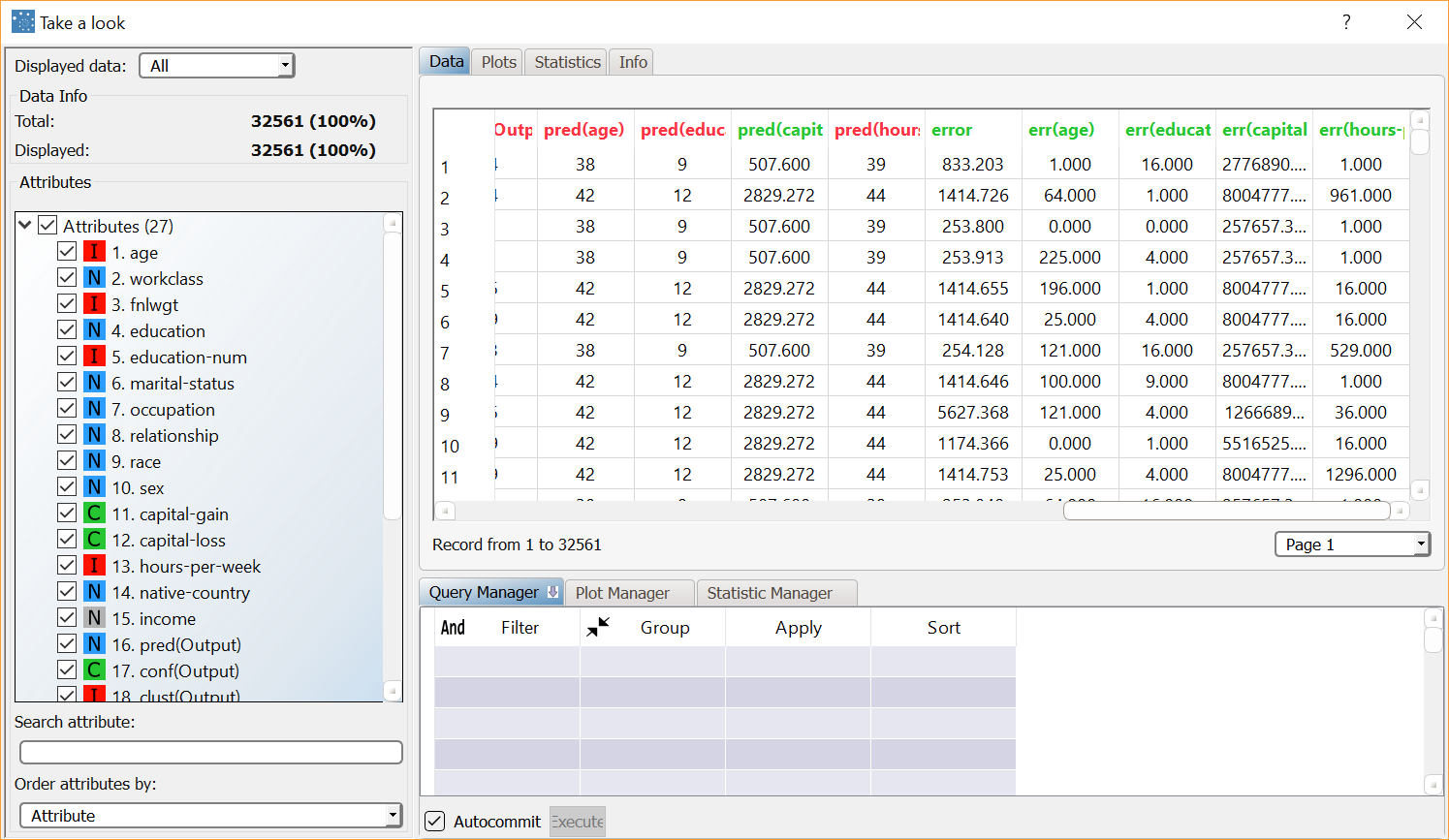
Add an Apply Model task to the process to create the index of the cluster to which each pattern in the training and in the test set belongs. This is obtained by finding the nearest centroid (according to the Distance and Normalization options selected in the Option panel of the Standard Clustering task. Compute the task leaving its default settings. To view the results, right-click the Apply Model task and select Take a look. 14 additional result variables have been added to the dataset. The first three result variables concern the cluster associated with the current pattern:
The subsequent five result variables report the values of the profile attributes for the centroid of the associated cluster: pred(age), pred(education-num), etc. The remaining six result variables concern the error generated when these values are employed as a forecast for the actual profile attributes of the pattern. In particular, the first of these result variables (error) provides the total error, whereas the others (err(age), err(education-name) etc.) provide the error for each attribute. Corresponding values for the patterns of the test set can be displayed by selecting Test set from the Displayed data drop-down list on the left. |